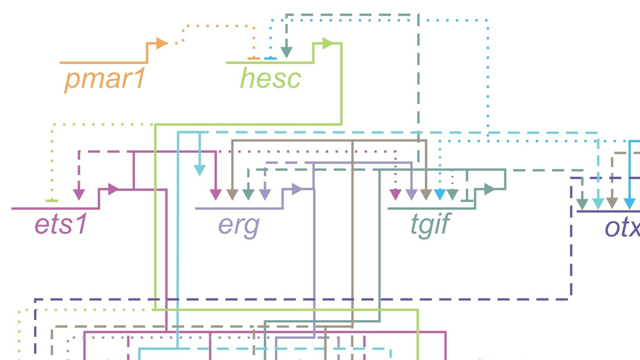
Our group has several broad interests that fall under the umbrella of “how genomes control cell fate decisions during development and how this evolves”. We think about development as embryonic and non embryonic (i.e. regeneration). We have ongoing projects in EvoDevo, including cooption, body plan evolution, and cis regulatory evolution, and in Regeneration, including, cell pluripotency, mechanobiology, and neurogenesis.
Our motto is to not be limited by approaches and technologies: therefore we work in molecular biology, genomics, biochemistry, microscopy, high throughput sequencing, embryology and computational biology. We’re very collaborative because we always want to find out how to do new things and we like working with diverse people. We also work at whatever scale of organization will best answer our questions, and thus we take very systems approaches as well as more reductionist approaches.
Our lab uses a range of echinoderm species as model systems which we house in our Marine Genomics Facility on the Mellon Institute. These organisms are especially well suited to high throughput analyses as we can readily generate and raise transgenic embryos and larvae. We work with multiple species of echinoderms, including sea urchins, Stronglyocentrotus purpuratus, Lytechinus variegatus, the sea sea star Patiria miniata and the sea cucumber Parastichopus parvimensis.
Our lab group runs Echinobase.org in which we work to provide and curate genome assemblies and genome feature annotations for community of researchers who use Echinoderms in their studies. Echinobase supported by NIH 1P41HD095831.