How do mechanical stresses in the local environment change cell fate?
We have a new funded collaborative project to measure the mechanical properties of cells undergoing pluripotency changes during sea star regeneration and to see how stresses on cells change their ability to differentiate. We plan to dig down into how nuclear stress effects chromatin and we’re working with Kris Dahl in Chemical Engineering. Project “Translating biological and mechanical properties from a natural in-vivo regenerating system for engineering human stem cells”. Funded by the DSF Scaife Charitable Foundation.
Here’s example of a sea star larvae healing after wounding.
Are there conserved mechanisms of regeneration?
Many animals have exceptional capacities for regeneration and like our sea star can reform a whole body even when bisected in half. We asked whether there were any similarities in the genes and mechanisms of regeneration in sea stars that are also used during regeneration of even distantly related species. Here is the summary from our latest paper: Published here
Below are highlights of some our our earlier work:
The GRN for endomesoderm specification in sea stars.
An important first step in making an embryo is for the cells to decide on fate choice between making the endomesoderm and the ectoderm. Our paper shows how in sea stars this choice depends on the gradients of beta-catenin which makes our paper the first empirical demonstration that beta catenin dose response can control cell fate choice in echinoderms.
McCauley BS, Akyar E, Saad HR, Hinman VF. Dose-dependent nuclear β-catenin response segregates endomesoderm along the sea star primary axis. Development. 2015 Jan 1;142(1):207-17. doi: 10.1242/dev.113043. PMID: 25516976
How Transcription Factors Can Evolve
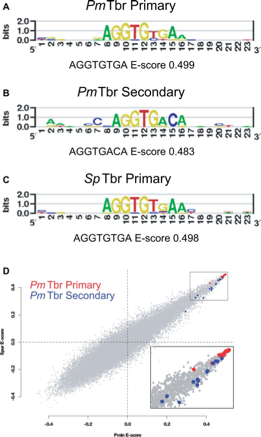 It is well established that a prominent source of evolutionary change in GRNs is of changes in regulatory DNA. A change in cis regulatory sequence allows a single node in the network to evolve, whereas when a transcription factor protein evolves, all of its target genes will be affected. Therefore it has been assumed that proteins are much more constrained. We were able to show however that Tbrain transcription factors are able to evolve a change in preference and affinity for their DNA binding domain. Sea urchin, sea stars and even mouse Tbrain genes share a highly similar high affinity binding site, but they have evolved a changed preference for an additional (secondary) low affinity binding site. This provides a previously undiscovered mechanism to evolve only a subset of their target genes. We are now working to understand:1. how these secondary sites function during development 2, Whether this mechanism of evolutionary change extends to other factors.
It is well established that a prominent source of evolutionary change in GRNs is of changes in regulatory DNA. A change in cis regulatory sequence allows a single node in the network to evolve, whereas when a transcription factor protein evolves, all of its target genes will be affected. Therefore it has been assumed that proteins are much more constrained. We were able to show however that Tbrain transcription factors are able to evolve a change in preference and affinity for their DNA binding domain. Sea urchin, sea stars and even mouse Tbrain genes share a highly similar high affinity binding site, but they have evolved a changed preference for an additional (secondary) low affinity binding site. This provides a previously undiscovered mechanism to evolve only a subset of their target genes. We are now working to understand:1. how these secondary sites function during development 2, Whether this mechanism of evolutionary change extends to other factors.
Cheatle Jarvela AM, Brubaker L, Vedenko A, Gupta A, Armitage BA, Bulyk ML, Hinman VF. Modular Evolution of DNA-Binding Preference of a Tbrain Transcription Factor Provides a Mechanism for Modifying Gene Regulatory Networks. Mol Biol Evol. 2014 Jul 12. pii: msu213
This work was featured as the Editors Choice in Science!
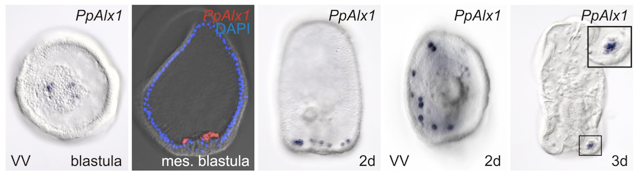
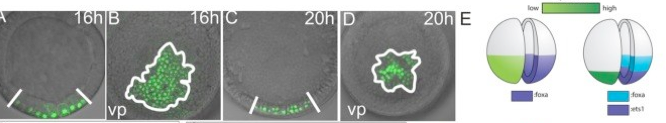
The GRN for neurogenesis in Echinoderm larvae.
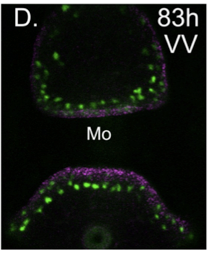 The different groups of echinoderm larvae have very different, and often quite complex patterns of neurons. This provides a great opportunity to understand how GRNs control development for precise patterning of highly patterned cell types, in this case a neuron. We are continuing this work to understand how patterning can evolve and how neuronal precursors make fate decisions that integrate specification and patterning.
The different groups of echinoderm larvae have very different, and often quite complex patterns of neurons. This provides a great opportunity to understand how GRNs control development for precise patterning of highly patterned cell types, in this case a neuron. We are continuing this work to understand how patterning can evolve and how neuronal precursors make fate decisions that integrate specification and patterning.
Yankura KA, Koechlein CS, Cryan AF, Cheatle A, Hinman VF (2013). Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc Natl Acad Sci U S A. 2013 May 6.
How GRNs evolve for novel morphologies
One of the driving questions in our group is how GRNs evolve for novel morphologies. This is important for understanding how the diversity of life has evolved and for understanding the ways that GRNs can change. Most of our analyses to date have focussed on GRN comparisons between sea urchins and sea stars because sea urchins have a novel cell lineage -the PMCs – which make the larval skeleton. Recently we have expanded this comparative work to include sea cucumber Parastichopus parvimensis as an experimental system. The embryos of this animal develop very similarly to sea stars, but have some characteristics of sea urchins. For instance we have discovered that these embryos have a PMC lineage that makes just a small larval spicule. The three way comparison between sea urchin, sea star and se cucumber embryos is allowing us to very particularly tease apart how GRNs can evolve for novel outcomes.
McCauley BS, Wright EP, Exner C, Kitazawa C, Hinman VF. (2012) Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. EvoDevo 3(1).