Research
The Impact of Genomic Plasticity on Ecological Success
Pneumococcal strains exhibit extensive genomic variability and plasticity. Thus, pneumococcal colonization must be understood from the perspective of a heterogeneous population of cells that represent a broad range of genomic variants. My lab investigates how strain-specific genomic content contributes to phenotypic diversity, gene regulation and ecological advantage.
One example of our studies on the relationship between accessory genes and ecological advantages focuses on the PMEN1 (pneumococcal molecular network clone 1). The PMEN1 is an archetype of genomic success: a pandemic and multi-drug resistant lineage that spread widely in the post-antibiotic era. Starting with the premise that genes unique to the PMEN1 lineage are an important component of this ecological success, we study strain-specific loci.
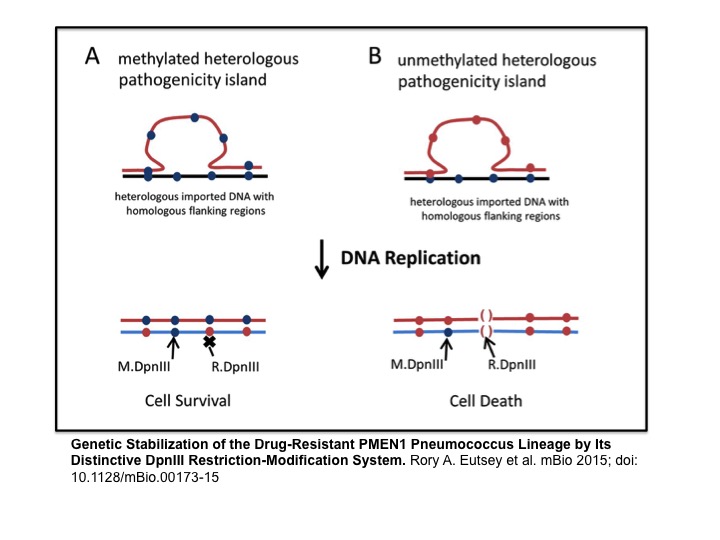
One feature of the PMEN1 lineage is that its strains are productive gene donors, but recalcitrant to uptake of DNA from strains in other lineages. Our studies of the DpnIII restriction-modification system, a locus unique to the PMEN1, showed that it limits recombination from non-PMEN1 strains while having no effect on recombination between PMEN1 strains (Eutsey et al, 2015). This is based on the dependence of DpnIII on the methylation state of the incoming DNA. Thus, our work suggests that DpnIII is the mechanism that allows the well-adapted PMEN1 lineage to increase genomic stability, rather than foster genomic plasticity.
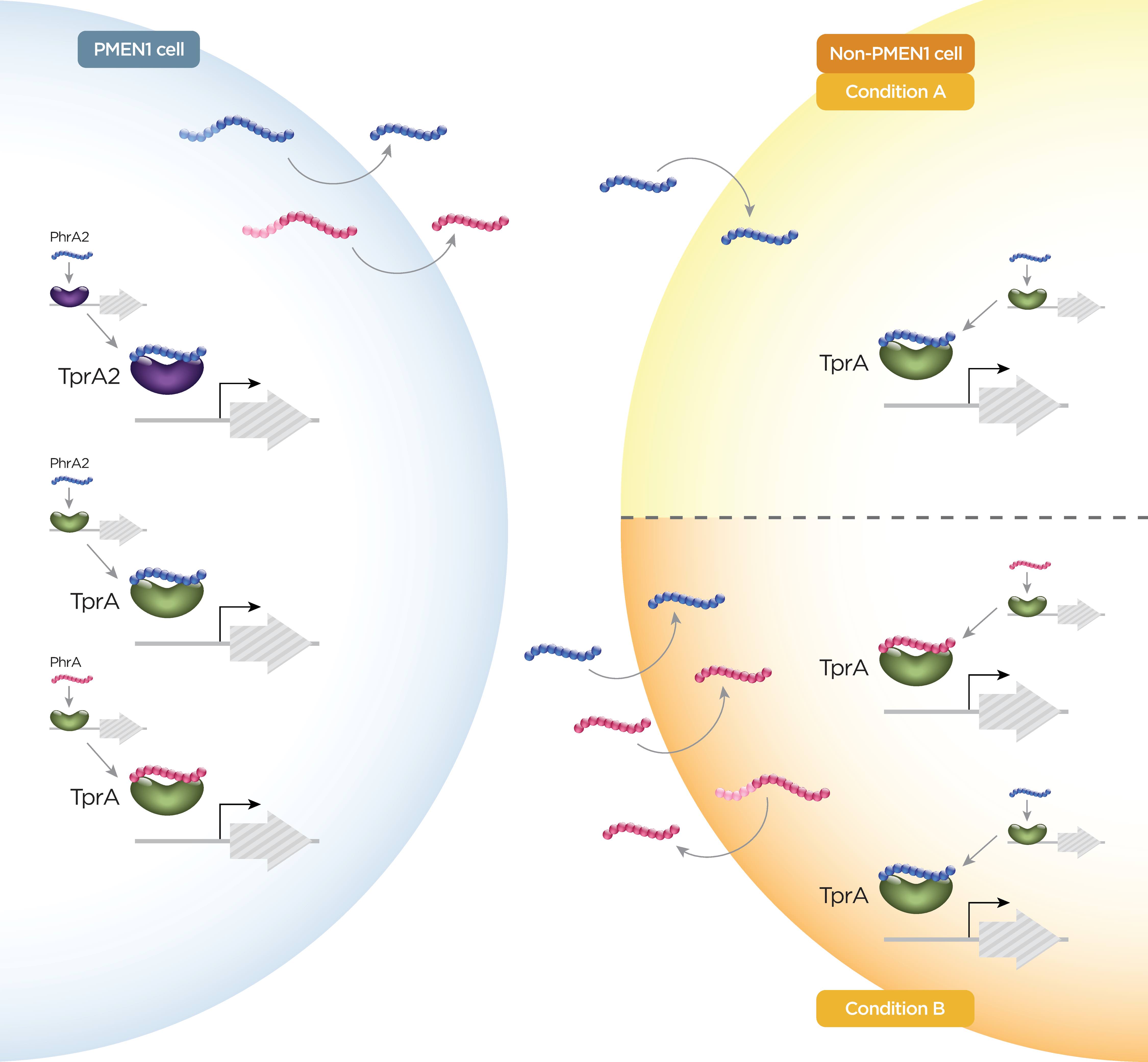
Another PMEN unique locus is the TprA2/PhrA2 system. Its characterization revealed one of the few known examples of inter-strain cross talk in pneumococcus. We showed that this system displays unidirectional cross-talk with a paralog system encoded in all strains (TprA/PhrA). This is especially relevant given that TprA/PhrA promotes virulence in multiple animal models of systemic disease and carriage. In this manner, our data suggest that a strain-specific molecule (PhrA2) activates a core virulence determinant (TrpA/PhrA). The ability of isolates from one lineage to manipulate the virulence of neighboring cells from a different lineage may have important clinical implications in that it suggests the virulence potential of a strain may depend on whether the infection is the result of a single-strain or multi-strain infection.